Introduction
OpenEdition (http://www.openedition.org) is composed of three different platforms dedicated to electronic resources in the humanities and social sciences, Revues.org (journal and book series), Hypotheses.org (research notebooks and scholarly blogs) and Calenda (social sciences calendar). Revues.org is the oldest French platform of academic online journals. It now offers more than 300 journals available in all of the disciplines of the humanities and social sciences. The online publication is made through the conversion of articles into XML TEI (http://www.tei-c.org) format and then into XHTML format and allows the viewing of the full text in web browsers. The bibliographical parts of articles on Revues.org are rather diverse and complicated. One main reason of this complexity is the diversity of source disciplines that makes various styles in reference formatting:
- References employ different journal stylesheets or do not follow any stylesheet,
- Labeling should be very precise: performance should be close (or better) those of a ‘human labeler’. In case this could not be achieved, the software should provide user with a confidence score. The quality of a digital library depends on the quality of metadata that allow field-based search, citation analysis, cross-linking and so on,
- References can be written in different languages,
- References contain some misspellings,
- Entries may be ambiguous (for example, searching for papers from ‘Williams Patrick’ in Google Scholar retrieve several papers: a) ‘Colonial discourse [...]’ published in 1996, b) ‘Mariage tsigane […]’ (gipsy wedding) published in 1984, c) ‘A randomized trial of group outpatient visits […]: the Cooperative Health Care Clinic’ published in 1997; we can assume that they correspond to different authors with the same name),
- Sometimes, one must refer to other references in order to complete them or else references are grouped together,
- References may be located anywhere in papers: in the body of the text, at the end of the paper and in footnotes.
Three new Training Corpora in DH Fields
In order to prepare a data set for the construction of a machine learning model (see below), which automatically estimates reference fields, we adopted a subset of TEI XML tags and annotated manually three different corpora according to three difficulty levels:
- Corpus level 1: the references are at the end of the article under a heading ”References”. Manual identification and annotation are relatively simpler than for the two next levels. Considering the diversity, 32 journals are randomly selected and 38 sample articles are taken. Total 715 bibliographic references were identified and annotated.
- Corpus level 2: the references are in notes and they are less formulaic compared to the ones of the corpus level 1. An important characteristic of the corpus level 2 is that it contains link informations between references. And because of the nature of note data, the corpus contains both bibliographical and non-bibliographical notes. We selected 41 journals from a stratified selection and we extracted 42 articles considering the diversity. We observed 1147 bibliographical references that were manually annotated and 385 non-bibliographical notes, which did not need any manual annotation.
- Corpus level 3: the references are in the body of articles. The identification and annotation are the most complicated. Even finding the beginning and end of bibliographic parts in a note is difficult. For example, we may accept a phrase citing a book title as reference even if its author is not found near by but in footnote. We selected 42 articles considering different properties of implicit reference in the body of the articles. From these selected articles, we identified and annotated 1043 references.
The following table shows some examples from these three corpora. In total, we annotated 3290 bibliographical references by using the tags presented on Table 1.
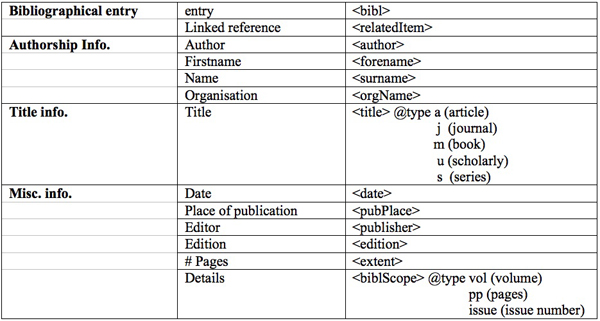
Table 1: Tags for annotating references
Machine Learning for Annotation
Most of the free softwares available on the Internet process pre-identified references against a set of predefined patterns. For example, cb2bib (http://www.molspaces.com/cb2bib/) recognizes reference styles of the publications of the American Chemical Society and of Science Direct but does not work on other styles without a costly adaptation. Some other softwares employ machine learning and numerical approaches by opposite to symbolic ones that require a large set of rules that are very hard to manage and that are not language independent. Day et al. (2005) cite the works of a) C.L. Giles et al. for the CiteSeer system (computer science literature) that achieves a 80% accuracy for author detection and 40% accuracy for page numbers (1997-1999), b) Seymore et al. that employ Hidden Markov Models (HMM) that learn generative models over input sequence and labeled sequence pairs to extract fields for the headers of computer science papers, c) Peng et al. that use Conditional Random Fields (CRF) (Lafferty et al. 2001) for labeling and extracting fields from research paper headers and citations. Other approaches employ discriminatively-trained classifiers (such SVM classifiers). Compared to HMM and SVM, CRF obtained better tagging performance (Peng & McCallum 2006). Some papers propose methods to disambiguate author citations (Han & Wu 2005; Torvik & Smalheiser 2009) or geographical identifiers (Volz et al. 2007). These ‘state of the art’ approaches seem to achieve good results but they proceed on limited size collections of scientific research papers only and they do not resolve all the difficulties we identified above. We choose Conditional Random Fields (CRFs) as method to tackle our problem of bibliographic references annotation on DH data (Kim et al. 2011). It is a type of machine learning technique applied to the labeling of sequential data. By definition, a discriminative model maximizes the conditional distribution of output given input features. So, any factors dependent only on input are not considered as modeling factors, instead they are treated as constant factors to output (Sutton & McCallum 2011). This aspect derives a key characteristic of CRFs, the ability to include a lot of input features in modeling. The conditional distribution of a linear-chain CRF for a set of label y given an input x is written as follows. The multiplication of all these conditional probabilities for input data is maximized during the iterative learning process.
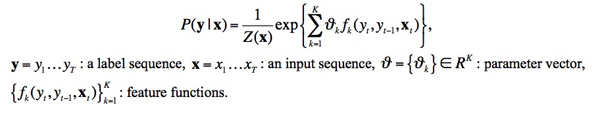
Experiments
We separately learn a CRF model for each corpus. Until now, we have conducted various experiments with the first and second corpora for the automatic annotation of reference fields. The standard on the effective local features and appropriate reference field labels are established during the construction of CRF model on the first corpus. This standard is equally applied to the second corpus but in this time, another automated process is necessary for the selection of target notes that include bibliographical information. Another machine learning technique, Support Vector Machine (Joachims 1999) is used for this note classification and we propose a new method, which mixes input, local and global features to well classify notes into bibliographical or non-bibliographical categories. The new feature generation method works well in terms of both classification accuracy and reference annotation accuracy. The features we selected for CRF are listed in Table 2.
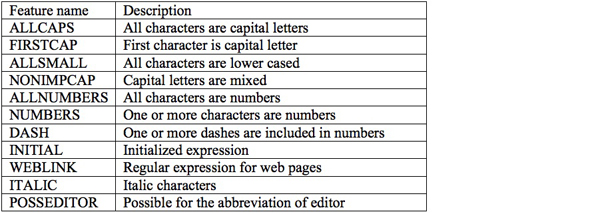
Table 2: Features employed for automatic annotation
A remarkable point is that we separately recognize authors and even their surname and forename. In traditional approaches, different authors in a reference are annotated as a field. Compared to the scientific research reference data used in the work of Peng and McCallum (2006), our corpus level 1 is much more diverse in terms of bibliographical reference formats. However, we have obtained a successful result in overall accuracy (90%) especially on surname, forename and title fields (92%, 90%, and 86% of precision respectively). They are somewhat less than the previous work (95% overall accuracy) but considering the difficulty of our corpus, current result is very encouraging.
The notes in the corpus level 2 are more difficult to treat and had not been studied in the previous works to the best of our knowledge. With our pre-selection process using a Support Vector Machine (SVM) classifier was effective to finally increase the automatic annotation result for corpus level 2. For the comparison, we constructed another CRF model on all notes without classification. Consequently, the CRF model learned with the classified notes using our approach outperforms the other model, especially on recall of three important fields, surname, forename, and title as in the following table. Bold character in the following table means that the corresponding model outperforms in the field with a statistical significance test (see http://bilbo.hypotheses.orghttp://bilbo.hypotheses.org for more information).
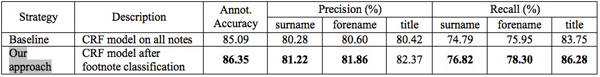
Table 3
Conclusion
We have constructed three different corpora of bibliographical reference in DH field. Then we have conducted a number of experiments to establish the standards of processing on the corpora. From the basic experiments on the first and second corpora, we continue to refine the automatic annotation via different techniques. Especially we develop an efficient methodology to incorporate incomplete external resources such as proper noun list to a CRF model. According to a trial experiment, the incorporation enhances automatic annotation performance on proper noun fields with corpus level 1. We will try the same method on the second corpus. Given that the annotation accuracies were not so high with the second corpus, we expect a more significance improvement. The integration of the note classification and annotation steps into a process is also expected to improve the efficiency of learning process. One of most important part will be the treatment of corpus level 3. We are now developing several approaches to deal with this corpus.
Acknowledgements
This work was supported by the Google Grant for Digital Humanities in 2011; and the French Agency for Scientific Research under CAAS project [ANR 2010 CORD 001 02].
References
Day, M.-Y., T.-H. Tsai, C.-L. Sung, C.-W. Lee, S. H. Wu, C. S. Ong, and W. L. Hsu (2005). A knowledge-based approach to citation extraction. Information Reuse and Integration. IRI -2005 IEEE International Conference, pp. 50-55.
Han, H., W. Xu, H. Zha, and C. Lee Giles (2005). A Hierarchical Naive Bayes Mixture Model for Name Disambiguation in Author Citations. Proceedings of SAC’05, ACM.
Joachims, T. (1999). Making large-scale support vector machine learning practical. Cambridge: MIT Press, pp. 169-184.
Kim, Y.-M., P. Bellot, E. Faath, and M. Dacos (2011). Automatic annotation of bibliographical reference in digital humanities books, articles and blogs. Proceedings of the CIKM 2011 BooksOnline11 Workshop.
Lafferty, J., A. McCallum, and F. Pereira (2001). Conditional Random Fields: Probabilistic Models for Segmenting and Labeling Sequence Data. International Conf. on Machine Learning (ICML’01).
Peng, F., and A. McCallum (2006). Information extraction from research papers using conditional random fields. Information Processing & Management 42(4): 963-979.
Torvik, V. I., and N. R. Smalheiser (2009). Author Name Disambiguation in MEDLINE. ACM Transaction on Knowledge Discovering in Data 3(3).
Volz, R., J. Kleb, and W. Mueller (2007). Towards ontology-based disambiguation of geographical identifiers. WWW int. conf., Canada.
Sutton, C., and A. McCallum (2011). An introduction to conditional random fields. Foundations and Trends in Machine Learning. To appear. http://arxiv.org/abs/1011.4088 (accessed 25 March 2012).

